Deposit Your Datasets in DV-IMPACT
The main purpose of DV-IMPACT is to serve as a central resource for large-scale disease variant impact assessment studies and to make these data available online in a uniform format that guarantees data integration and interoperability between different datasets. This page lists the requirements for datasets deposition to DV-IMPACT . The dataset should comply with the DV-IMPACT standard data model (Table 1) and has the structure (or can be processed to be in the structure) that fits DV-IMPACT (Figure 1).
| Data Items | Attributes | Type | Description |
| Mutations | Protein ID | Text | Standard protein identifier e.g. UniProt or Ensemble IDs (e.g. P04637 or ENSP00000269305). |
| Mutation Type | Text | e.g. Substitution, Insertion, Deletion (as defined by mutation source database e.g. COSMIC). | |
| Mutation Description | Text | Should be in the standard form used by the major database and preceded by "p.". For instance, a substitution mutations will be "p.A1#A2", where A1 and A2 are the wildtype and the mutation amino acids, respectively, and # is the mutation position (e.g. p.P210R). | |
| Disease Name | Text | As mentioned in the original paper can also be abbreviated (e.g. AML for Acute Myeloid Leukemia). | |
| Mutation Source | Text | Database from where the mutations obtained (e.g. COSMIC, ICGC or TCGA). | |
| [O] Mutation ID | Text | The mutation identifier in the mutation source for reference. | |
| [O] Genomic Mutation Description | Text | Should be in the standard form used by the major database and preceded by "g." for genomic DNA. For instance, a substitution mutations in the DNA will be "g.N1#N2", where N1 and N2 are the wildtype and the mutation nucleotides, respectively, and # is the mutation position (e.g. g.A550C). | |
| [O] Mutation Recurrence | Number | How many times that same mutation was observed in the sample cohort it was identified in. | |
| [O] Mutation Samples | Number | Number of samples with the mutations to the total tested sample X/Y. | |
| Domains | Domain ID | Text | Unique identifier for each domain instance. |
| Domain InterPro ID | Text | The domain type identifier from InterPro Database. | |
| Protein ID | Text | Standard protein identifier of the protein where this domain is found. | |
| Domain Type | Text | The domain type as in from InterPro (e.g. SH2, SH3, WW, PDZ, Tyrosine-protein kinases). | |
| Domain Name | Text | A human readable name to identify the domain (e.g. ProteinName_# where ProteinName is the name of the protein where this domain is found and # is the order of the domains, in case of multiple domain proteins). | |
| Domain Coordinates* | Number | The domain start and end positions in the protein. | |
| [O] Other Proteins IDs | Text | Other proteins that have the same exact domain sequence. | |
| Proteins | Protein ID | Text | Standard protein identifier e.g. UniProt or Ensemble IDs (e.g. P04637 or ENSP00000269305). |
| Protein sequences | Text | The full protein sequence or the database and version information so that others can get the exact sequences was used in the predictions. | |
| [O] Protein Name | Text | Protein full name (e.g. Heat shock protein 90 for HSP90). | |
| [O] Gene Symbol | Text | Standard gene symbol (e.g. HGNC gene symbol). | |
| [O] Protein Description | Text | Additional information about the protein (e.g. other names, function or subcellular localization). | |
| Interactions | Domain Protein ID | Text | The protein identifier of the protein of the domain as in the Domains data item. |
| Peptide Protein ID | Text | The protein identifier of the protein of the peptide as in the Proteins data item. | |
| Domain ID | Text | The domain instance identifier as in the Domains data item. | |
| Wildtype Score | Number | Score/evaluation of wildtype predicted interaction e.g. p-value. | |
| Variant Score | Number | Score/evaluation of variant predicted interaction e.g. p-value. | |
| Binding Start* | Number | Peptide's start position.** | |
| Binding End* | Number | Peptide's end position. | |
| [O] Supporting Information | Text | Any information used to support the predicted PPI or rewiring event e.g. gene/protein expression, subcellular localization, disorder, surface accessibility or sequence conservation. | |
| [O]PWMs | PWM Name | Text | A name to identify the PWM e.g. DomainName_# where DomainName is the name of the domain which this PWM corresponds to and # is the order of the PWM, in case of multiple specificity domains. |
| PWM File | Text | Contains peptide sequences used to generate the PWM (in FASTA format) or the PWMs data. | |
| Domain ID | Text | The domain instance identifier as in the Domains data item. | |
| [O] Properties | Number | Any additional properties for the PWM e.g. score cut off or p-values. |
[O] Optional data item or attribute.
* Alternately, the peptide sequence can be provided.
** all sequence coordinates start from 1.
Table 1. The data items and their required and optional attributes according to DV-IMPACT data standard.
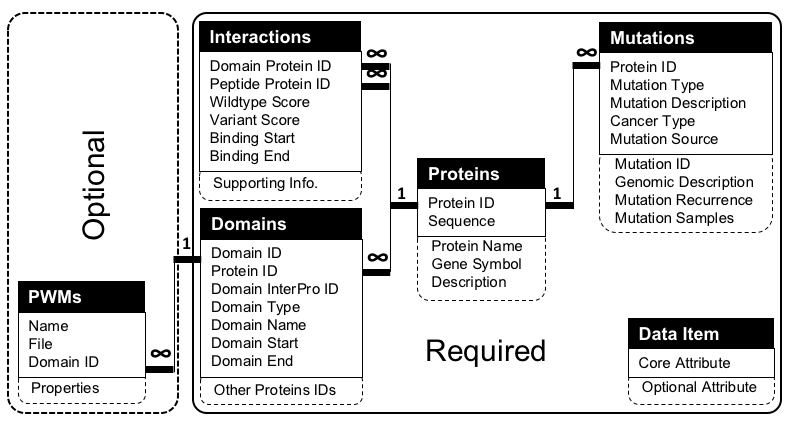
Figure 1. Relationships between data items of DV-IMPACT standard data model for disease variants impact assessment. The relationships between the attributes correspond to the relationships between the tables in DV-IMPACT .
If you want to deposit your data into DV-IMPACT , please contact us.


